
Multivariate analysis of mass spectrometry imaging data | contact@mvatools.com


Dear user
A new version of simsMVA is out and it includes a number of new features. This will be a rather long list but we would like to highlight all major updates:
You can now find on the top menu 18 different example datasets of various different structures. This includes 4 examples of analytical techniques other than SIMS. This list of examples is as follows:
Datasets can now be saved into .mat files and subsequently loaded in any other new tab of the same kind (Spectra, Profiles, Images or 3D). No more stitching the same set of patches every time or waiting to load a long 3D dataset!

Additionally to a series of bug fixes in the 3D data loader, the 3D visualiser and the 3D overlay tools were redesigned to allow more customisation (they are also now applicable to both the original data AND MVA results!).
The Z-correction tool has also been improved. The user can now pre-process the data and select whether to base the correction on a substrate or topographical feature



It is now possible to convert a 3D dataset into 2D (imaging) or 1D (depth profiling) data. The converted dataset will be loaded on a new tab, maintaining its “hyperspectral” aspect. This is an extremely useful tool that was developed out of need (no other software can do that).

It had been long overdue, but it is finally possible to add peak labels in modes of analysis other than “Spectra”. A button has been added next to the variable selector in each mode.

And it is possible to change markers’ size!

Thank you very much for your support and feedback.
For this tutorial you will be using the example data “Adhesive” from the examples menu in simsMVA. The data consists of secondary ion maps of the surface of a packaging adhesive. The aim is to produce the following figure:

Where the left-hand side contains an overlay of three NMF components intensities and the right-hand side contains their characteristic spectra.
To load the example dataset, click on “Examples” on the top menu and select “Imaging (Adhesive)”, then click on the new tab created.

Pre-process the data by normalising all maps by total ion counts and Poisson-scaling. Choose a more appropriate colour map that would help to identify whether everything is OK with the pre-processed data.

For this example, NMF was performed in a subset of only 3% of the pixels. This is done by selecting “Sample 3%” in the subsampling drop-down menu, then click the “NMF” ![]() button and set the input parameters:
button and set the input parameters:
A new tab is then created with the NMF results. Click on it.
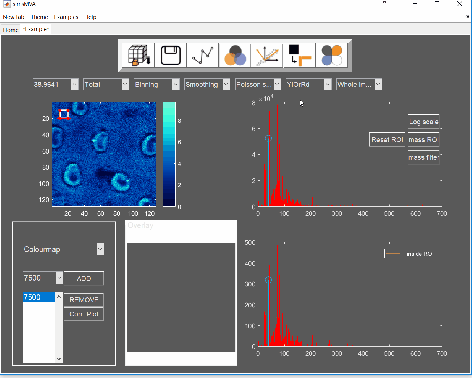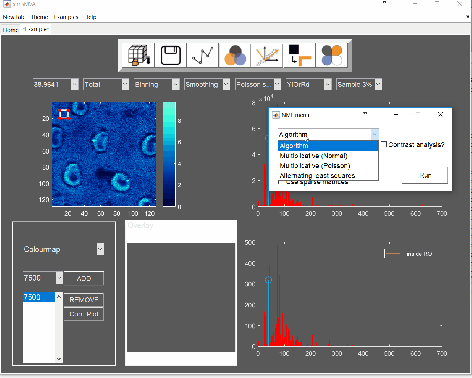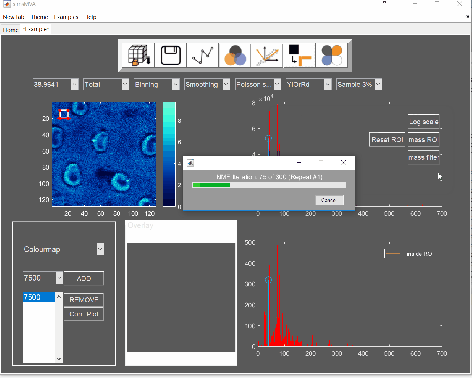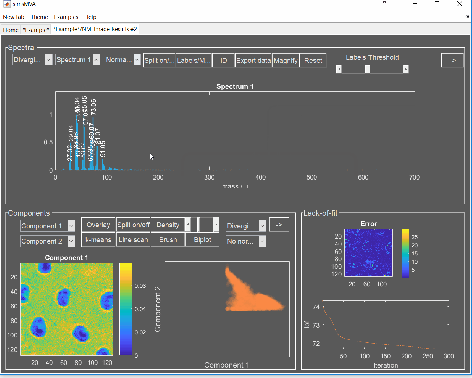
Now we have the data needed for the overlay. To produce it, click the “Overlay” button on the “Components” panel. If you intend to print it or include it in a white-background slide, change simsMVA colour scheme by clicking “Theme” on the top menu bar and selecting a white background theme such as “Almond Milk”.
The “Overlay” window consists of three panels: “Control”, “Overlay” and “Components”. On the “Control” panel, the “Add” buttons allow you to add individual components to the overlay and the coloured button next to it sets their respective colours. For this image, a base colourmap (“hsv”), without much tweaking, was enough to produce good results. In cases where there are more than three components, you may want to tweak the colours of a base colourmap in order to enhance contrast.

Note that the plots on the right-hand side are connected, so if you use the magnifying lens ![]() to zoom in any of the plots, all three plots will show the same x-scale.
to zoom in any of the plots, all three plots will show the same x-scale.

Once you are happy with the colour and sizes, you can export the figure by clicking “File->Export setup…”, select appropriate rendering and export it to your desired compression format.

A bit of background
Non-negative matrix factorisation (NMF)1,2 is a non-supervised machine learning method that seeks to reduce the dimensionality of a dataset down to a few “pure” components. For the application of NMF to ToF-SIMS images, the data is unfolded into a Matrix A containing samples (pixels) in rows and variables (spectral channels) in columns. The method then finds an approximate factorisation A ≈ WH into non-negative factors W and H. H is interpreted as the characteristic spectra of components and W is interpreted as the “concentration” distribution of the components over the samples. The number of components is defined by the inner dimension of the product WH, as shown in the figure below:
