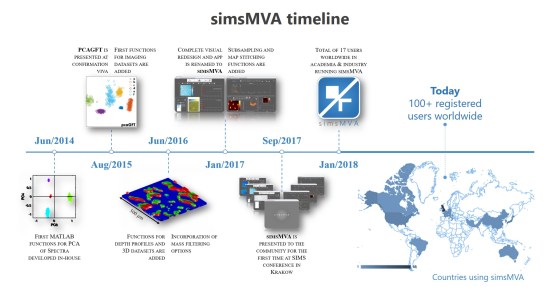
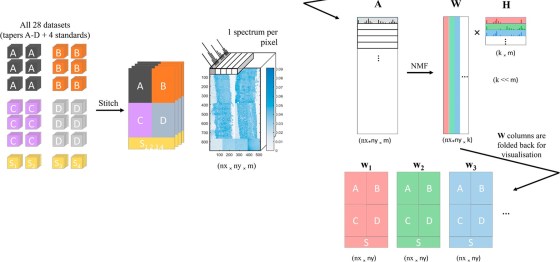
More info here: https://www.npl.co.uk/events/101st-iuvsta-workshop
The 101st IUVSTA workshop on high performance SIMS instrumentation and machine learning / artificial intelligence methods for complex data will be held between 19 and 24 of May 2024 in Maresias, Brazil.